Note
Go to the end to download the full example code
Explained variance (joined figure)¶
This example will show the explained variance from a principal component analysis as a function of the number of principal components considered. Here we join four different plots together.
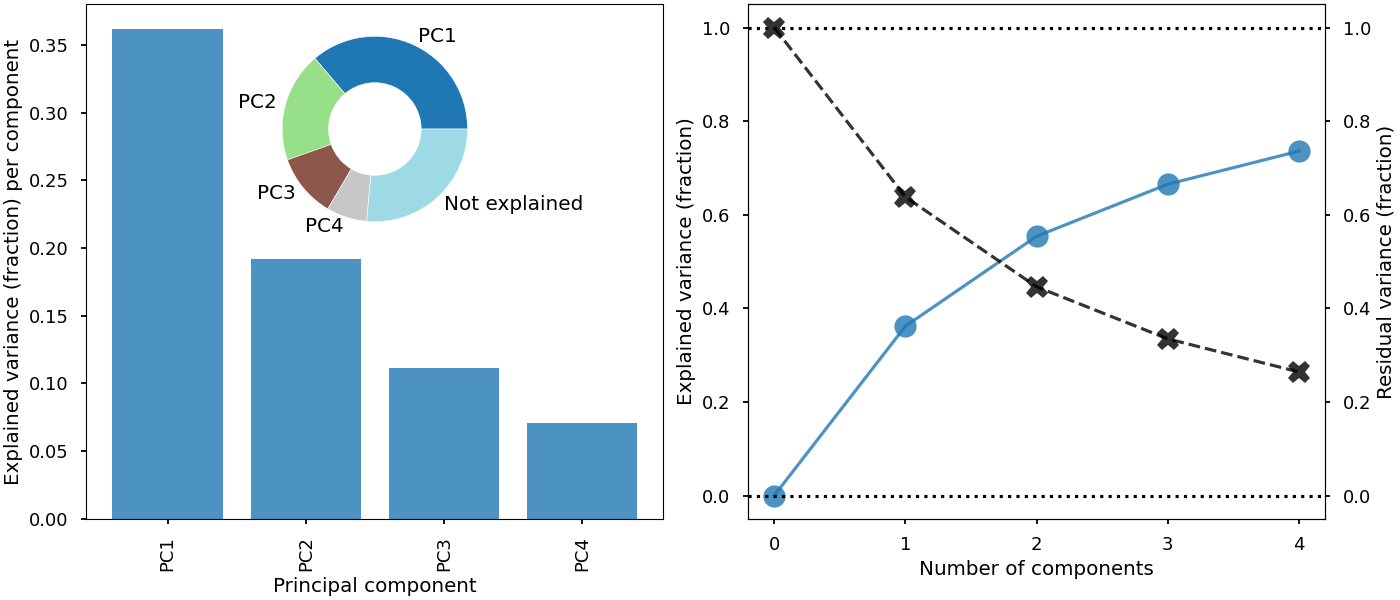
from matplotlib import pyplot as plt
from mpl_toolkits.axes_grid1.inset_locator import inset_axes
import pandas as pd
from sklearn.datasets import load_wine
from sklearn.preprocessing import scale
from sklearn.decomposition import PCA
from psynlig import (
pca_explained_variance,
pca_residual_variance,
pca_explained_variance_bar,
pca_explained_variance_pie
)
plt.style.use('seaborn-talk')
data_set = load_wine()
data = pd.DataFrame(data_set['data'], columns=data_set['feature_names'])
data = scale(data)
pca = PCA(n_components=4)
pca.fit_transform(data)
fig, (ax1, ax2) = plt.subplots(
nrows=1, ncols=2, figsize=(14, 6), constrained_layout=True
)
pca_explained_variance_bar(pca, axi=ax1, alpha=0.8)
pca_explained_variance(pca, axi=ax2, marker='o', markersize=16, alpha=0.8)
ax4 = ax2.twinx()
pca_residual_variance(
pca,
ax4,
marker='X',
markersize=16,
alpha=0.8,
color='black',
linestyle='--'
)
ax3 = inset_axes(ax1, width='45%', height='45%', loc=9)
pca_explained_variance_pie(pca, axi=ax3, cmap='tab20')
plt.show()
Total running time of the script: ( 0 minutes 0.480 seconds)